Introduction
The previous tutorials focus on the individual functions in the package, but do little to show how to integrate them into common workflows to develop spatial products for sampling. Here we show a minimal example of how a curator may load occurrence data for a species, apply a couple sampling approaches to these data, and save the results for use. We will use two species native to the Southwestern United States & Mexico for this example, eventually narrowing our focus to discuss only one for the sake of brevity.
While we only have two species in the example, the code is setup to accomodate as many species as an analyst desires to process simultaneously.
Data prep
You will need to install rgbif to follow along with this
example. rgbif is an amazing package maintained by ROpenSci
to acess the Global Biodiversity Information Facility database from
within R. Note that a (free, and esay to get), GBIF profile is required
for larger data downloads, but for this example all you need is the R
package.
library(safeHavens)
library(sf) # spatial data
library(dplyr) # general data handling
library(tidyr) # general data handling
library(purrr) # mapping functions across lists.
library(ggplot2) ## for maps
library(rgbif) # species occurrence data.
library(spData) # example cartography dataUsing rgbif we’ll download occurrence data for a couple
species, just so we have the code set up for mapping
through multiple species in a more realistic manner.
## small subset of useful columns for example
cols = c('decimalLatitude', 'decimalLongitude', 'dateIdentified', 'species', 'acceptedScientificName', 'datasetName',
'coordinateUncertaintyInMeters', 'basisOfRecord', 'institutionCode', 'catalogNumber')
## download species data using scientificName, can use keys and lookup tables for automating many taxa.
cymu <- rgbif::occ_search(scientificName = "Vesper multinervatus", limit = 1000)
### check to see what CRS are in here, these days usually standardized to wgs84 (epsg:4326)
table( cymu[['data']]['geodeticDatum'])
geodeticDatum
WGS84
675
## subset the data to relevant columns
cymu_cols <- cymu[['data']][,cols]
## repeat this again so a second set of data are on hand
bowa <- rgbif::occ_search(scientificName = "Bouteloua warnockii", limit = 1000)
bowa_cols <- bowa[['data']][,cols]
## contrived multispecies example.
spp <- bind_rows(bowa_cols, cymu_cols) |>
drop_na(decimalLatitude, decimalLongitude) |> # any missing coords need dropped.
st_as_sf(coords = c( 'decimalLongitude', 'decimalLatitude'), crs = 4326, remove = F)
rm(cymu, bowa, cols, cymu_cols, bowa_cols)Take a quick look at the data to see if there are any clear errors. One points location is incorrect, it’s latitude is great - we remove it below.
western_states <- spData::us_states |> ## for making a quick basemap.
dplyr::filter(REGION == 'West' & ! NAME %in% c('Montana', 'Washington', 'Idaho', 'Oregon', 'Wyoming') |
NAME %in% c('Oklahoma', 'Texas', 'Kansas')) |>
dplyr::select(NAME, geometry) |>
st_transform(4326)
ggplot() +
geom_sf(data = western_states) +
geom_sf(data = spp, aes(color = species, shape = species)) +
theme_void() +
theme(legend.position = 'bottom')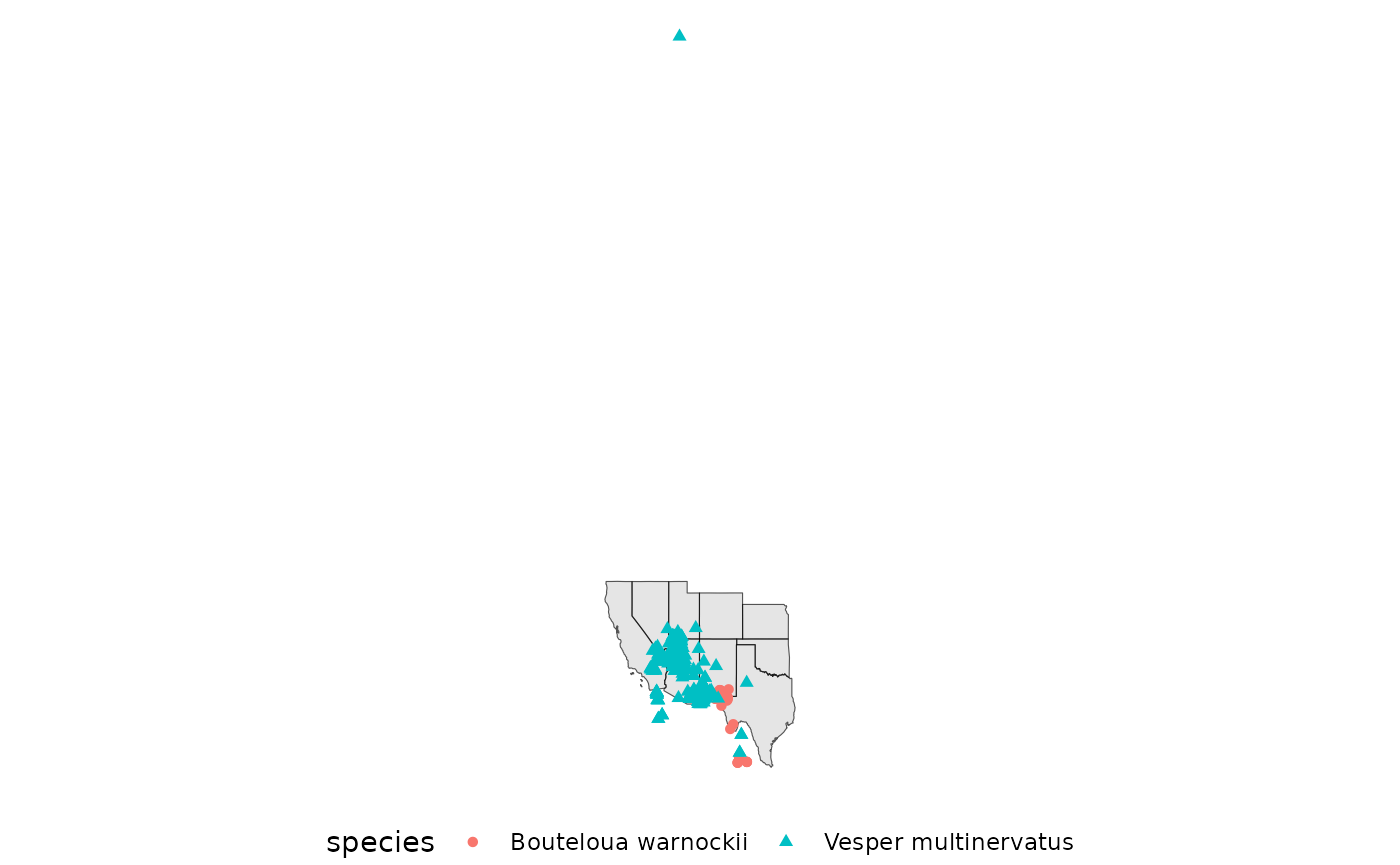
## check the outlying record.
arrange(spp, by = decimalLatitude, desc=FALSE) |>
head(5)
Simple feature collection with 5 features and 10 fields
Geometry type: POINT
Dimension: XY
Bounding box: xmin: -102.8461 ymin: 26.2436 xmax: -101.343 ymax: 26.30833
Geodetic CRS: WGS 84
[38;5;246m# A tibble: 5 × 11
[39m
decimalLatitude decimalLongitude dateIdentified species acceptedScientificName
[3m
[38;5;246m<dbl>
[39m
[23m
[3m
[38;5;246m<dbl>
[39m
[23m
[3m
[38;5;246m<chr>
[39m
[23m
[3m
[38;5;246m<chr>
[39m
[23m
[3m
[38;5;246m<chr>
[39m
[23m
[38;5;250m1
[39m 26.2 -
[31m103
[39m
[31m.
[39m
[31mNA
[39m Boutel… Bouteloua warnockii G…
[38;5;250m2
[39m 26.2 -
[31m103
[39m
[31m.
[39m
[31mNA
[39m Boutel… Bouteloua warnockii G…
[38;5;250m3
[39m 26.2 -
[31m103
[39m
[31m.
[39m
[31mNA
[39m Boutel… Bouteloua warnockii G…
[38;5;250m4
[39m 26.3 -
[31m101
[39m
[31m.
[39m
[31mNA
[39m Boutel… Bouteloua warnockii G…
[38;5;250m5
[39m 26.3 -
[31m101
[39m
[31m.
[39m
[31mNA
[39m Boutel… Bouteloua warnockii G…
[38;5;246m# ℹ 6 more variables: datasetName <chr>, coordinateUncertaintyInMeters <dbl>,
[39m
[38;5;246m# basisOfRecord <chr>, institutionCode <chr>, catalogNumber <chr>,
[39m
[38;5;246m# geometry <POINT [°]>
[39m
## remove it based on it's latitude.
spp <- filter(spp, decimalLatitude <= 40)A few tools exist for quickly cleaning GBIF data, gatoRs
is a recently published great choice for this.
Map the data again, and keep the ggplot around as a basemap for the rest of vignette.
bb <- st_transform(spp, 5070) |>
st_buffer(100000) |>
st_transform(4326) |>
st_bbox()
western_states <- st_crop(western_states, bb)
base <- ggplot() +
geom_sf(data = western_states, color = 'white') +
geom_sf(data = spp, aes(color = species, fill = species)) +
theme_void() +
theme(legend.position = 'bottom')
base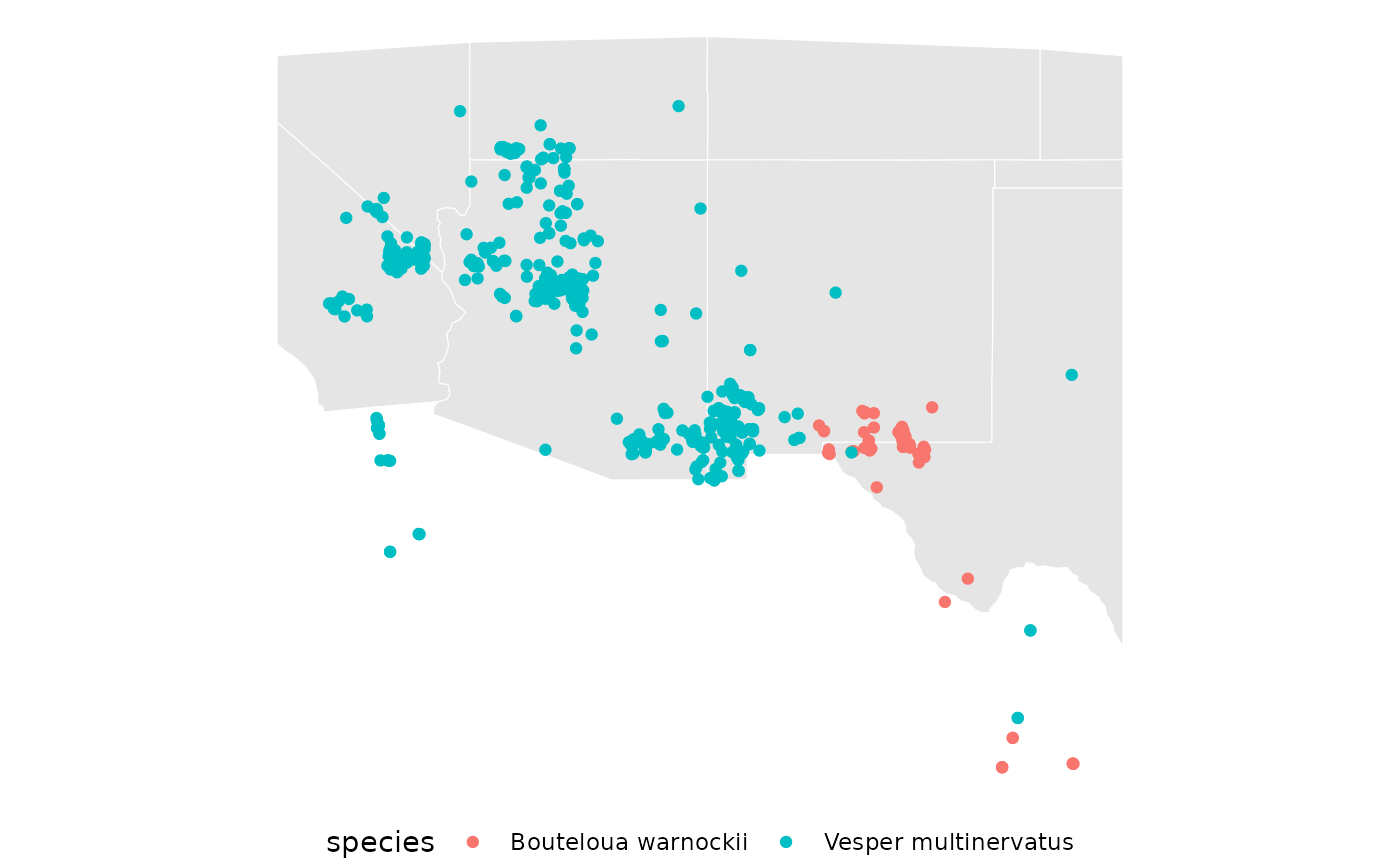
using safeHavens
Now we will use the safeHavens functionality with our set-up environment and data from GBIF.
Create species ranges
We will showcase two methods of generating the species range
geometries, which are used by most of the packages functions. Both of
these rely on the st_concave_hull function from
sf. st_concave_hull, has a ratio parameter
which can control the elasticity of hulls, with the ratio = 1 creating a
convex hull (akin the st_convex_hull function, also in
sf), and ratio = 0.0, creating a true concave hull. Results
below a ratio of 0.4 look too narrowly reduced for both of these
species.
sppL <- split(spp, f = spp$species)
concavities <- function(x, d, rat){
species <- x[['species']][1]
out <- st_transform(x, 5070) |>
st_buffer(dist = d) |>
st_union() |>
st_concave_hull(ratio = rat) |>
st_sf() |>
rename(geometry = 1) |>
mutate(species, .before = geometry)
}
spp_concave <- sppL |>
purrr::map(~ concavities(.x, d = 20000, rat = 0.4))
spp_convex <- sppL |>
purrr::map(~ concavities(.x, d = 20000, rat = 1.0))
rm(concavities)Visualize the concave hulls.
base +
geom_sf(data = spp_concave[[1]], aes(fill = species), alpha = 0.2) +
geom_sf(data = spp_concave[[2]], aes(fill = species), alpha = 0.2) 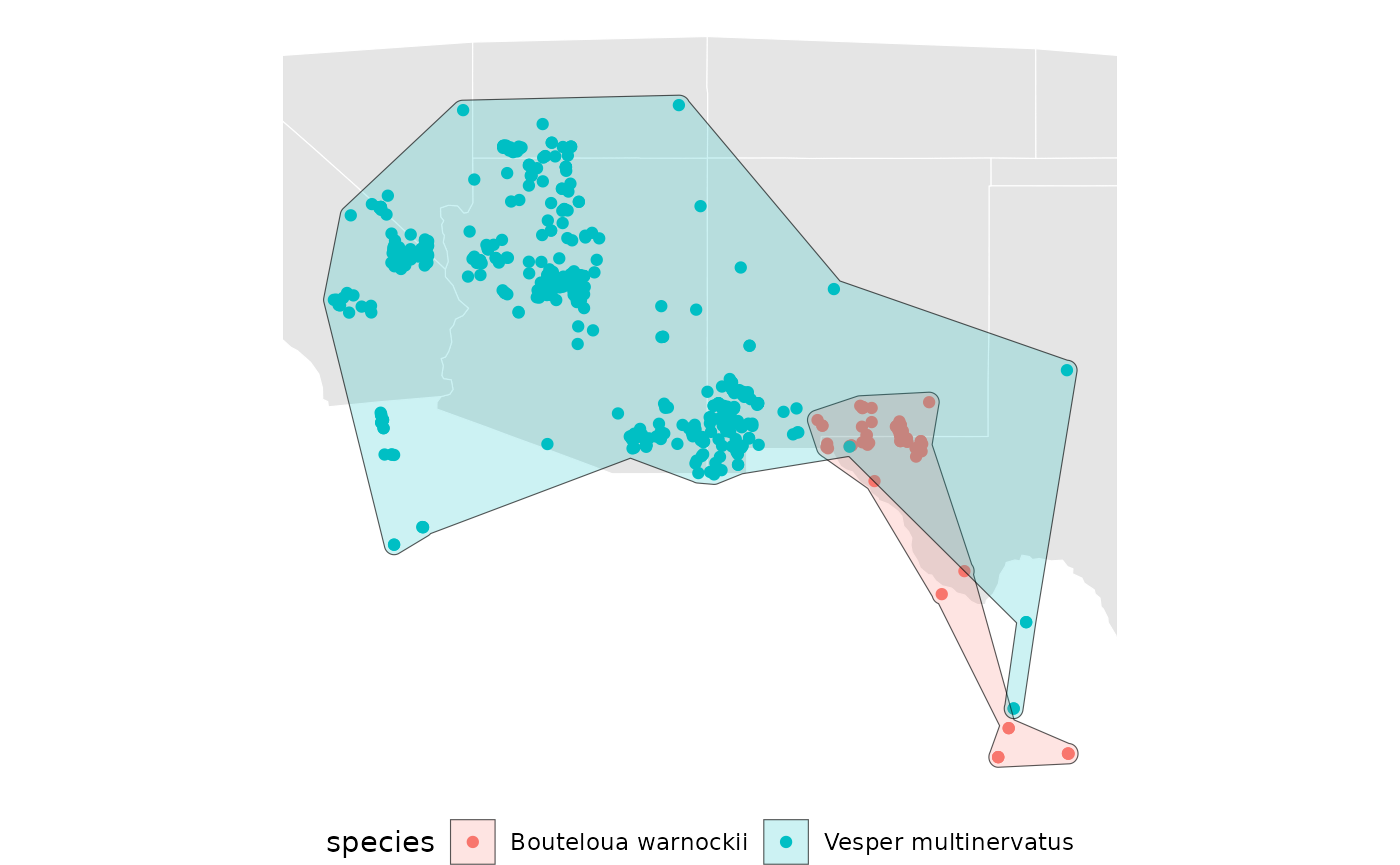
Visualize the convex hulls.
base +
geom_sf(data = spp_convex[[1]], aes(fill = species), alpha = 0.2) +
geom_sf(data = spp_convex[[2]], aes(fill = species), alpha = 0.2) 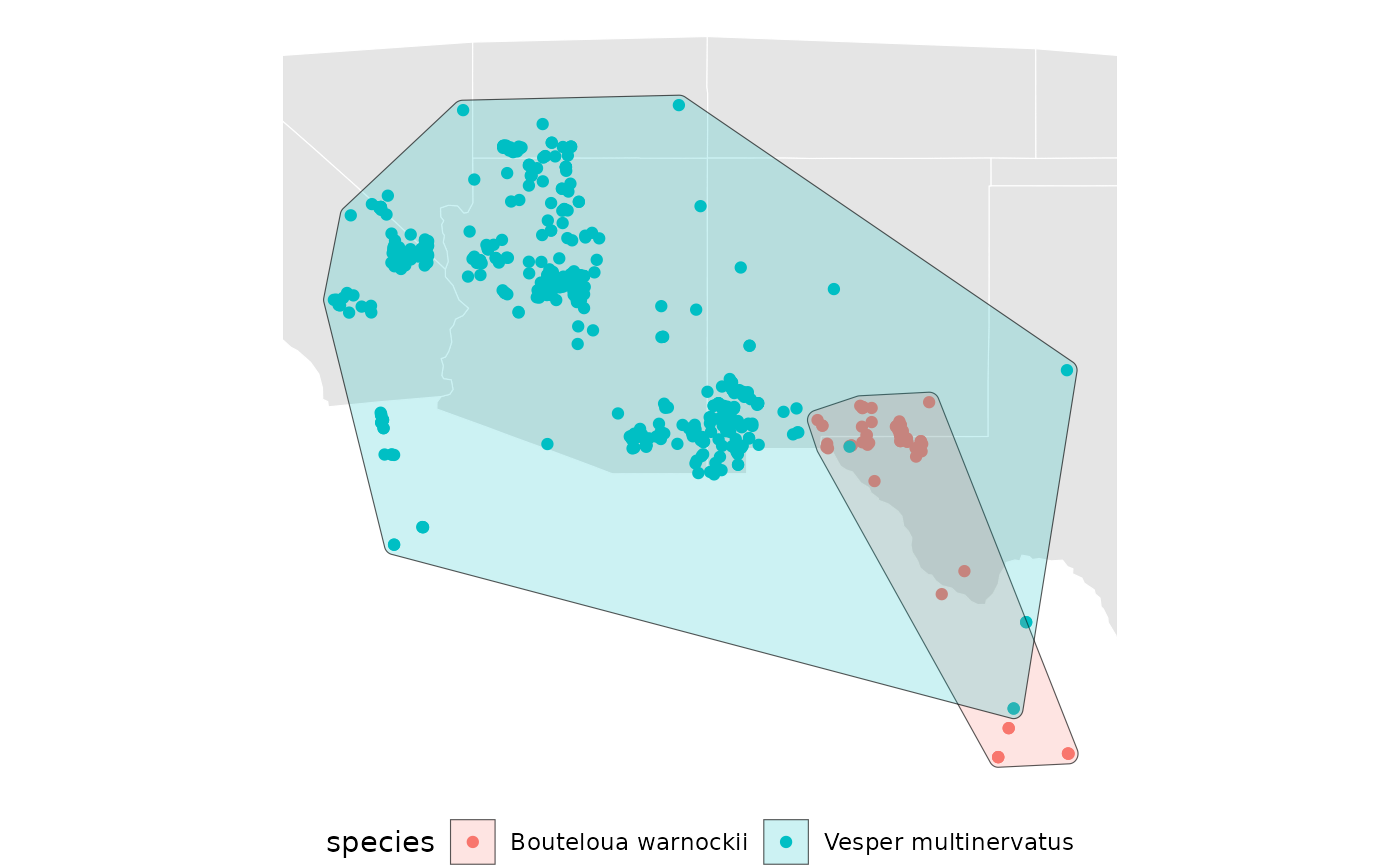
rm(spp_convex)For the sake of the example we will only use the concave ranges, with ratio 0.4, going forward.
Perform sampling for the species ranges
First we will perform equal area sampling with a relatively small amount of points and repetitions.
eas <- spp_concave |>
purrr::map(~ EqualAreaSample(.x, n = 10, pts = 250, planar_proj = 5070, reps = 25))
base +
geom_sf(data = eas[[2]][['Geometry']], aes(fill = factor(ID)), alpha = 0.2)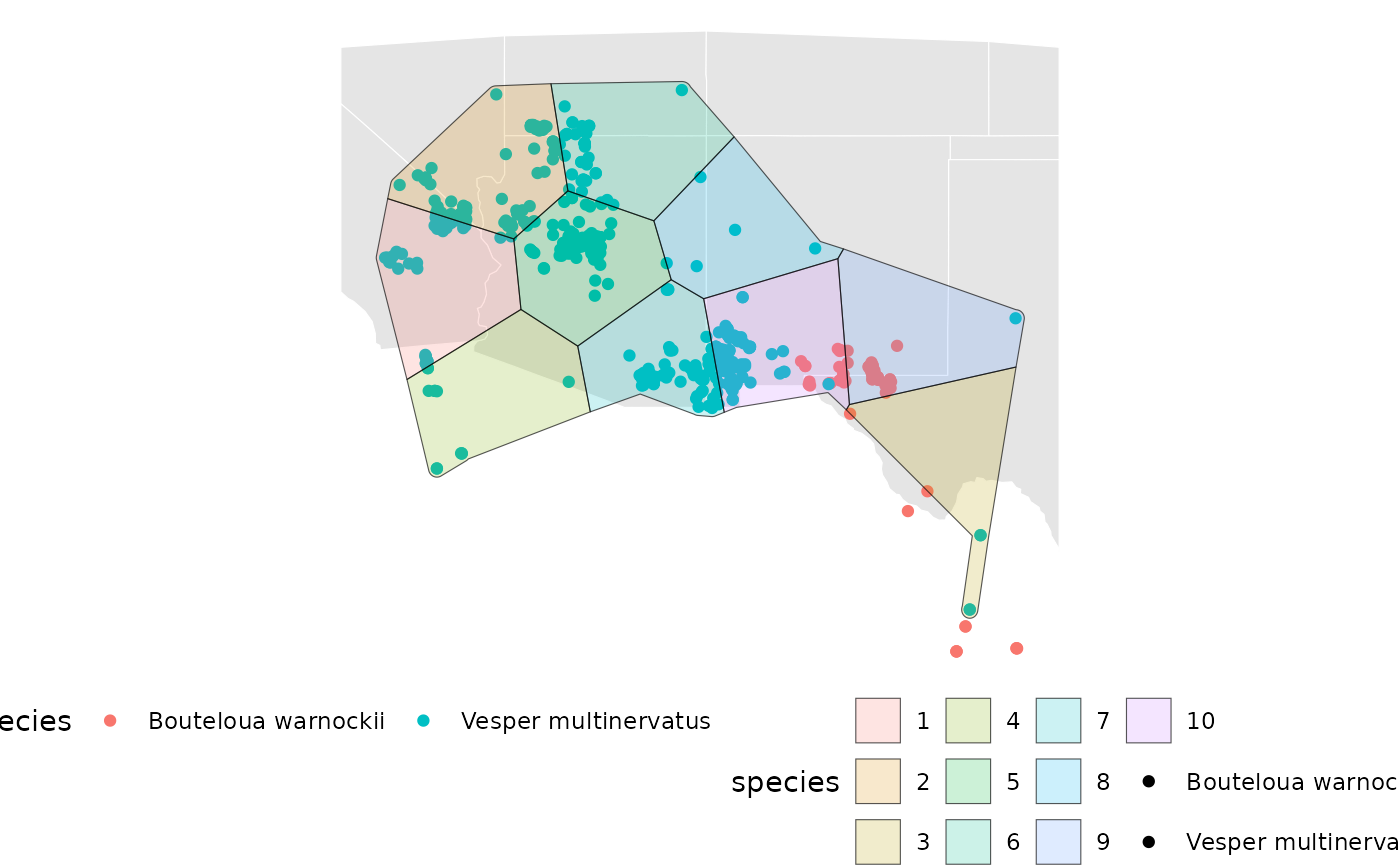
The plot above shows only the results for Vesper multinervatus.
Below we perform isolation by distance (IBD) based sampling.
# create an arbitrary template for example - best to do this over your real range of all species collections
# so that it can be recycled across species.
template <- terra::rast(terra::ext(bb), crs = terra::crs(spp), resolution = c(0.1, 0.1))
terra::values(template) <- 0
# the species range now gets 'burned' into the raster template.
spp_concave <- purrr::map(spp_concave, \(x) {
st_as_sf(x) |>
mutate(Range = 1, .before = geometry) |>
st_transform(4326) |>
terra::rasterize(template, field = 'Range')
})
## the actual sampling happens here within `map`
ibd_samples <- spp_concave |>
purrr::map(~ IBDBasedSample(.x, n = 10, fixedClusters = FALSE, template = template, planar_proj = 5070))
base + ## visualize for a single taxon.
geom_sf(data = ibd_samples[[1]][['Geometry']], aes(fill = factor(ID)), alpha = 0.2)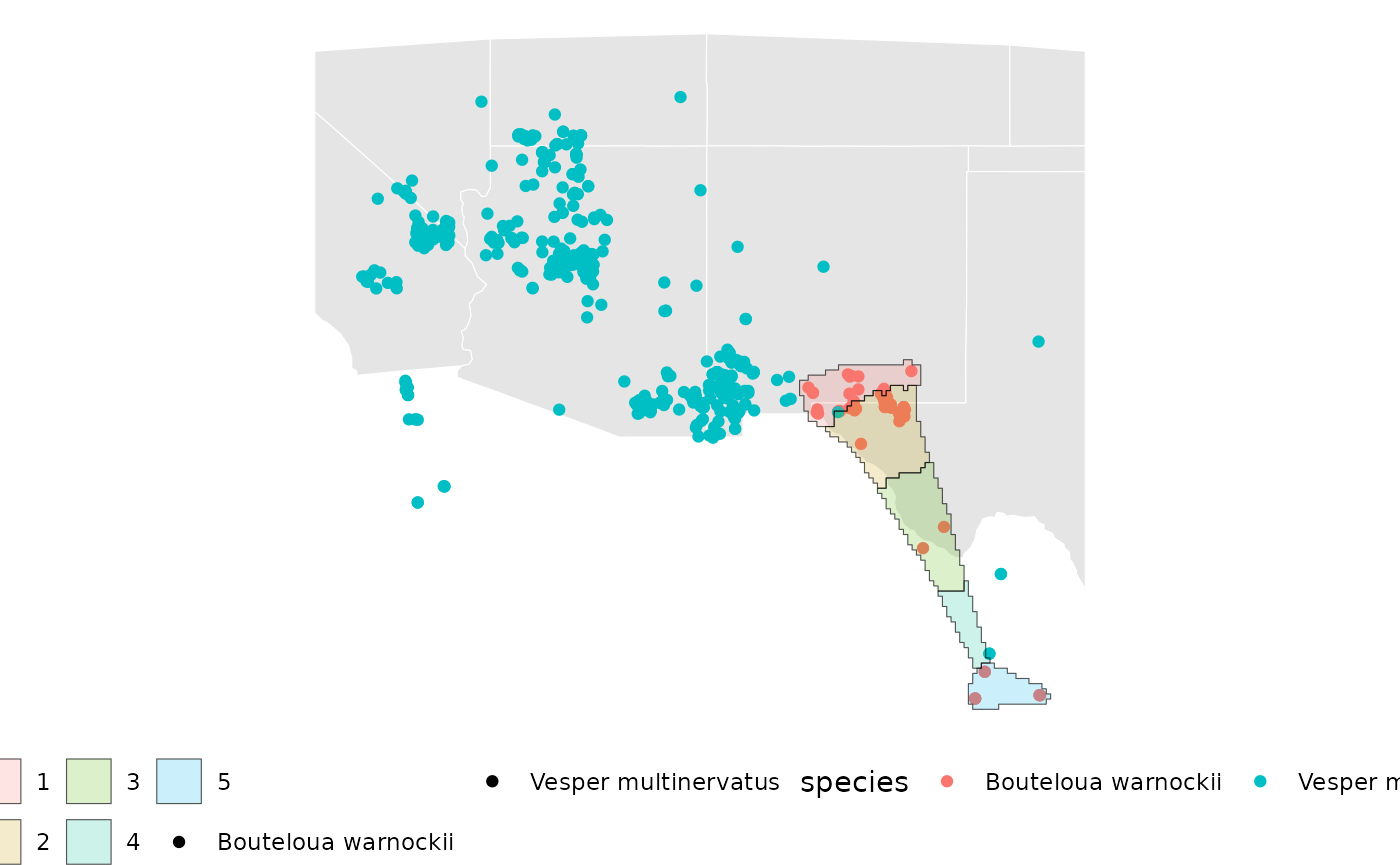
rm(spp_concave)The above plot for the isolation-by-distance sampling shows the individual sample areas for Bouteloua warnockii.
prioritize sample areas
In addition to creating spatial geometries that can be used to guide
sampling efforts, safeHavens can also help provide
visualize guidance on general areas where germplasm can be
sampled from to try and maximize distance between the samples. Please
note that while safeHavens can offer suggestions
of where to sample, and where should be prioritized these
suggestions may not align with reality - so consider these rules of
thumbs! I have made, a couple hundred large native seed collections, and
coordinated a couple hundred more, for many years - and argue that
beggers cannot be choosers. Hence basing deliverable metrics purely
around data like this can be difficult - some degree of reporting
autonomy must be maintained.
The function PrioritizeSample offers two levels of
prioritization. The more coarse level will suggest a relative order that
the sample areas should be sampled in, from 1:n; the function seeks to
minimize the variance ( a related measure) between each sample as more
samples are collected. In effect it tries to stratify the sampling
across the species range. The second part of the function is almost a
‘heamap’ which can be used to show concentric rings around the
geographic center of each sample unit. Keeping the number of rings low
allows for some pragmatic communication of more desirable/less desirable
portions of the range to ideally collect in.
ibd_samples_priority <- ibd_samples %>%
purrr::map(~ st_transform(.x$Geometry, 5070)) |>
purrr::map(~ list(PrioritizeSample(.x, n_breaks = 3)))
base +
geom_sf(data = ibd_samples_priority[[2]][[1]][['Geometry']], aes(fill = factor(Level)), alpha = 0.2)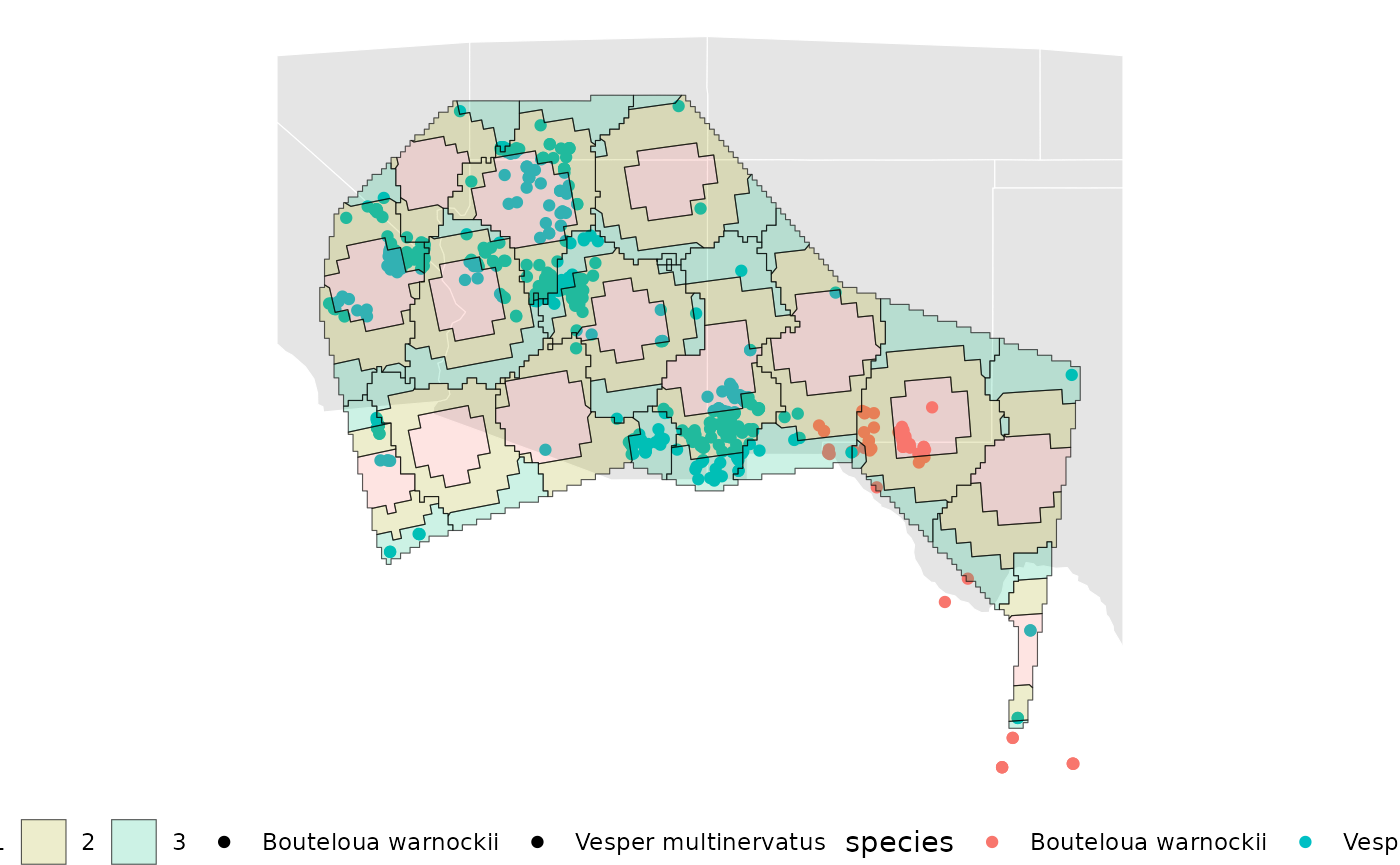
The map above shows a possible general order to guide the prioritization of individual sample areas, and simplified visuals of where within sample areas to attempt to target.
wrapping up
Upon completion of using safeHavens results should be
written to individual geopackages for long term storage. A strong
benefit of a geopackage is it’s ability to hold multiple geometry types
(polygons, points, rasters, etc.), which can sometimes be lost when they
are kept in separate directories.
Here we show how we would write out data for a range of species.
## create a directory to hold the outputs
p2Collections <- file.path('~', 'Documents', 'WorkedExample_Output')
dir.create(p2Collections, showWarnings = FALSE)
## we will only save the template raster once since it is recycled across taxa.
dir.create(file.path(p2Collections, 'IBD_raster_template'), showWarnings = FALSE)
terra::writeRaster(template,
filename = file.path(p2Collections, 'IBD_raster_template', 'IBD_template.tif'), overwrite = FALSE)
## save each species as a unique geopackage.
for(i in seq_along(sppL)){
fp = file.path(p2Collections, paste0(gsub(' ', '_', sppL[[i]]$species[1]), '.gpkg'))
### GBIF occurrence points
st_write(sppL[[i]], dsn = fp, layer = 'occurrence_points', quiet = TRUE)
### results of equal area sampling
st_write(eas[[i]]$Geometry, dsn = fp, layer = 'equal_area_samples', quiet = TRUE, append = TRUE)
### ibd based sample (note can be reconstruced from the hulls of ibd samples priority)
st_write(ibd_samples[[i]]$Geometry, dsn = fp, layer = 'ibd_samples', quiet = TRUE, append = TRUE)
### prioritized information for the IBD samples.
st_write(ibd_samples_priority[[i]][[1]]$Geometry, dsn = fp, layer = 'ibd_sampling_priority', quiet = TRUE, append = TRUE)
message(format(object.size(fp), standard = "IEC", units = "MiB", digits = 4))
}Warning in rm(spp, sppL, eas, ibd_samples, ibd_samples_priority, base,
template, : object 'p2Collections' not found
Warning in rm(spp, sppL, eas, ibd_samples, ibd_samples_priority, base,
template, : object 'fp' not found
Warning in rm(spp, sppL, eas, ibd_samples, ibd_samples_priority, base,
template, : object 'i' not found